Community Tip - Your Friends List is a way to easily have access to the community members that you interact with the most! X
- Subscribe to RSS Feed
- Mark Topic as New
- Mark Topic as Read
- Float this Topic for Current User
- Bookmark
- Subscribe
- Mute
- Printer Friendly Page
Avoid filterNaN error if there is no NaN
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Avoid filterNaN error if there is no NaN
I have various data sets I'm performing some analysis on. Sometimes they have blanks, sometimes not. I can easily filter out the blanks with filterNaN. The problem is if there are no blanks the filterNaN errors out. So, I have to rewrite the statements for each column as to whether I need to filter or not. I'd like one command that works whether I have blanks or not. What are my options? Here's a simple example:
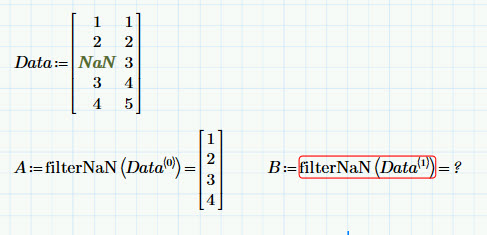
Here, I want to return B = [1 2 3 4 5]^T
Prime 3.1 (and is this a bug?)
Thanks,
Solved! Go to Solution.
Accepted Solutions
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
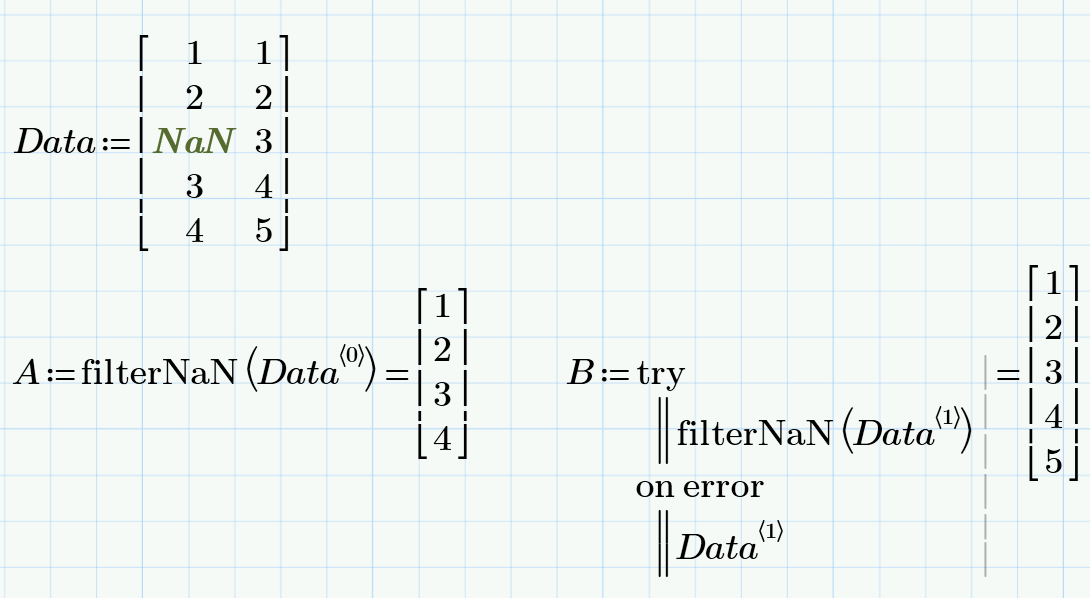
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Write a short program with "try..on error"
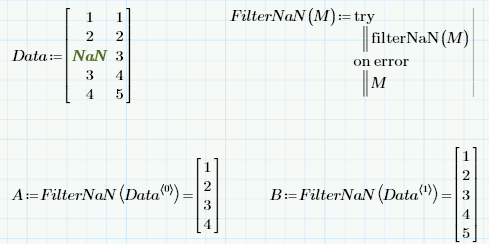
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
That's a very elegant solution.
But what if the data is paired--the rows are time sequential for example. The solution presented does not maintain that pairing; the resulting vectors are different lengths.
The attached (much less elegant) program will maintain the row correlation of a data set while stripping out any rows with missing data.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Excellent work as always Fred. My immediate problem did not require keeping data aligned in rows, as I was just making histograms and didn't want to eliminate rows needlessly. However, I was toying with the idea that I'd need your solution going forward since each row of data actually aligns with a test unit. So, I'll definitely be saving this one. Thanks!
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Fred Kohlhepp wrote:
That's a very elegant solution.
But what if the data is paired--the rows are time sequential for example. The solution presented does not maintain that pairing; the resulting vectors are different lengths.
I may be missing the point, but filterNaN works on vectors AND matrices exactly the way you would like it to be and consequently Marks modification does so, too.
It CAN be applied to very column of a matrix separately, but if you apply it on the full matrix. every row with even just one NaN is deleted.
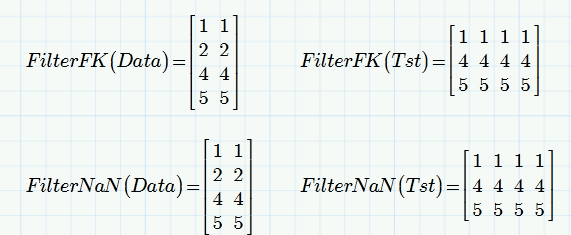
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
You're right! (as usual.) I had not seen that.
Fred
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
Fred,
I can't open your file, but I think the simplest way to maintain rows of data would be to filter before separating columns:
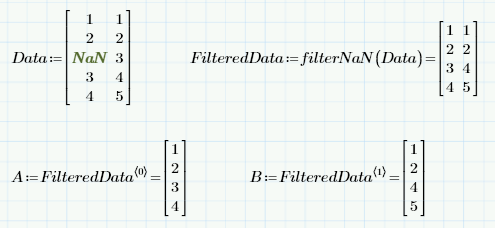
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
File is Prime 3.0.
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator
I only have 1.0 on this machine (purchased for v15).
- Mark as New
- Bookmark
- Subscribe
- Mute
- Subscribe to RSS Feed
- Permalink
- Notify Moderator





